Background: The delineating point of the end of the brainstem and beginning of the spinal cord is known as the cervicomedullary junction (CMJ). This point is defined by the decussation of the pyramidal tracts. Abnormal configuration and location of the CMJ have both been implicated in disease processes such as Chiari malformation. Unfortunately, the CMJ is not directly visualized on contemporary imaging techniques. Diffusion tensor imaging (DTI) has given us the ability to directly visualize white matter tracts, but suffers from difficulties with visualizing crossing fibers. Many advanced techniques for visualizing crossing fibers utilize substantially long imaging times or non-clinical magnet strengths making clinical applicability limited. Objective: This study investigates the use of generalized q-sampling imaging with diffusion decomposition on standard DTI acquisitions at 3 Tesla to demonstrate the pyramidal decussation. Methods: Three differing DTI protocols were analyzed in vivo with scan times of 17 minutes, 10 minutes, and 5.5 minutes. Data was processed with standard DTI post-processing, as well as generalized q-sampling imaging with diffusion decomposition. Results: The results of the study show that the pyramidal decussation can be reliably visualized using generalized q-sampling imaging and diffusion decomposition with scan times as low at 10 minutes. Conclusion: Utilizing generalized q-sampling post-processing, the pyramidal decussation can be reliably visualized using clinically feasible DTI sequences with scan times as low as 10 minutes. Since the data can be obtained from a clinically feasible acquisition, further investigations into abnormalities of the cervicomedullary junction should be practical utilizing this method.
The corticospinal tract (CST) is a long fiber pathway, which is essential for voluntary motor function. The tract originates in the motor cortex and travels through the corona radiata, internal capsule, and cerebral peduncle en route to the medulla. In the lower medullary segment, approximately 80% of fibers cross to the contralateral side and continue in the spinal cord as the lateral corticospinal tract. The point of crossing is termed the pyramidal decussation and defines the end of the brainstem and beginning of the spinal cord (cervicomedullary junction).
The cervicomedullary junction has important implications both in a diagnostic role and in surgical planning. Several studies have linked an abnormal cervicomedullary junction to Chiari malformations. These include both an abnormal structural appearance and abnormal location [1] [2] [3]. Of particular interest is the high association of an abnormally low-positioned cervicomedullary junction in Chiari malformations independent of cerebellar tonsillar position [3]. The pyramidal decussation can also be abnormal or absent in conditions such as Dandy Walker syndrome, Joubert’s syndrome, and horizontal gaze palsy and progressive scoliosis (HGPS) [4]. The inability to directly identify the cervicomedullary junction, as defined by the decussation of the pyramids, on conventional imaging has led to difficulty with further in vivo studies of this phenomenon.
Recent progress in MRI diffusion tensor imaging (DTI) has given us the tools to further unlock the complex architecture and function of the human brain. Unfortunately, a major limitation with DTI has been the difficulty with resolving crossing fibers [5]. Newer acquisition techniques such as high angular resolution diffusion imaging (HARDI) have improved our ability to detect crossing fibers [6]. Combination with model-based methods of reconstruction, such as spherical harmonic decomposition [7], has also led to improved fiber resolution. These methods, however, still have shown limitations with visualizing crossing fibers [8] [9].

More advanced model-free techniques have shown significantly improved results with crossing fiber resolution. Examples include q-space imaging [10] and diffusion spectrum imaging (DSI) [11]. The methodology and limitations are beyond the scope of this article; however, a major limitation is the inherently long acquisition times. A previously published study successfully showing the pyramidal decussation was obtained in ex vivo specimens at 11.7 Tesla [12]. Since current clinical practice only allows magnet strengths to 3 Tesla, this has severe limitations in clinical implementation. New methods of image processing, such as generalized q-sampling imaging method (GQI) [9], have improved the ability to resolve crossing fibers using standard DTI data sets. A proposed method of obtaining ODF by diffusion decomposition has recently been published and shows promising results [8].
The aim of our study is to accurately demonstrate the decussation of the pyramids utilizing clinically feasible imaging acquisition with the assistance of the aforementioned data processing techniques. This would allow further investigations into the role of the pyramidal decussation in certain pathologic conditions.
| Subject 1 | Subject 2 | Subject 3 | |
|---|---|---|---|
| b-value | 1000 | 1000 | 1000 |
| Voxel Size | 2 x 2 x2 mm | 2 x 2 x 2 mm | 2 x 2 x 2 mm |
| Diffusion Directions | 48 | 48 | 30 |
| No. of Acquisitions | 2 | 1 | 1 |
| Acceleration Factor | 4 | 4 | 2 |
| Scan Time | 17 mins. | 10 mins. | 5.5 mins. |
Diffusion tensor imaging acquired in 3 healthy volunteers without any known neurological disease was analyzed. The subjects were scanned using a 3 Tesla MRI (Vantage Titan, Toshiba, Tokyo, Japan). Diffusion data was acquired using a spin-echo based echo-planar imaging (EPI) diffusion pulse sequence with motion-sensitizing gradients and a 32-channel head coil. Three separate acquisitions were obtained with differing protocols. For all scans, the field of view = 256 x 256 mm, matrix size = 128 x 128, slice thickness = 2 mm (no gap), voxel size = 2 x 2 x 2 mm, and b-value of 1000 s/mm2.
For Subject 1, the scan was obtained at 48 directions with a TR/TE = 10000/82 ms, average number = 2, and an acceleration factor (SPEEDER) = 4 resulting in a scan time of 17 minutes. The scan in Subject 2 was obtained at 48 diffusion directions with a TR/TE = 12000/82 ms, average number = 1, and an acceleration factor of 4 resulting in a scan time of 10 minutes. Subject 3 was scanned at 30 diffusion directions with a TR/TE = 10000/82 ms, average number = 1, and an acceleration factor of 2 resulting in a scan time of 5.5 minutes. The scan parameters are summarized in Table 1.

A detailed anatomic survey was also performed using a 3D GRE T1-weighted sequence acquired in the sagittal plane. The slice thickness = 1.2 mm (no gap), TR/TE = 7/4 ms, TI = 900 ms, flip angle = 8, and average number = 1. The FOV = 256 x 256 mm and matrix size = 208 x 208.
Image processing was performed with DSI Studio (//dsi-studio.labsolver.org). Initially, standard DTI reconstruction was performed. Regions of interest were placed to isolate the corticospinal tract through the midbrain and pons. Tractography was performed with an FA threshold = 0.15, max angle = 65°, step size = 1 mm, and length constraint = 25-450 mm. The starting orientation for seeding was set to explore all orientations utilizing a subvoxel seeding strategy with trilinear orientation. The tracking was performed utilizing the Runge-Kutta method (RK4). A total of 400 tracts were obtained.
Next, the source images were processed using GQI. The diffusion sampling length ratio was calculated with r-squared weighting at 1.20. ODF Sharpening was performed utilizing Decomposition with a decomposition fraction (ε) = 0.05 and maximum fiber population (m) = 8. The number of fibers resolved per voxel was 6 and with a 6-fold ODF Tessellation.
Tractography was performed in DSI Studio. Regions of interest were placed to isolate the corticospinal tract through the midbrain and pons. The QA threshold = 0.205, max angle = 65°, step size = 1 mm, smoothing = 0.30, and length constraint from 25-450 mm. The starting orientation for seeding was set to explore all orientations utilizing a subvoxel seeding strategy with trilinear orientation. The tracking was performed utilizing the Runge-Kutta method (RK4). A total of 200 tracts were obtained.

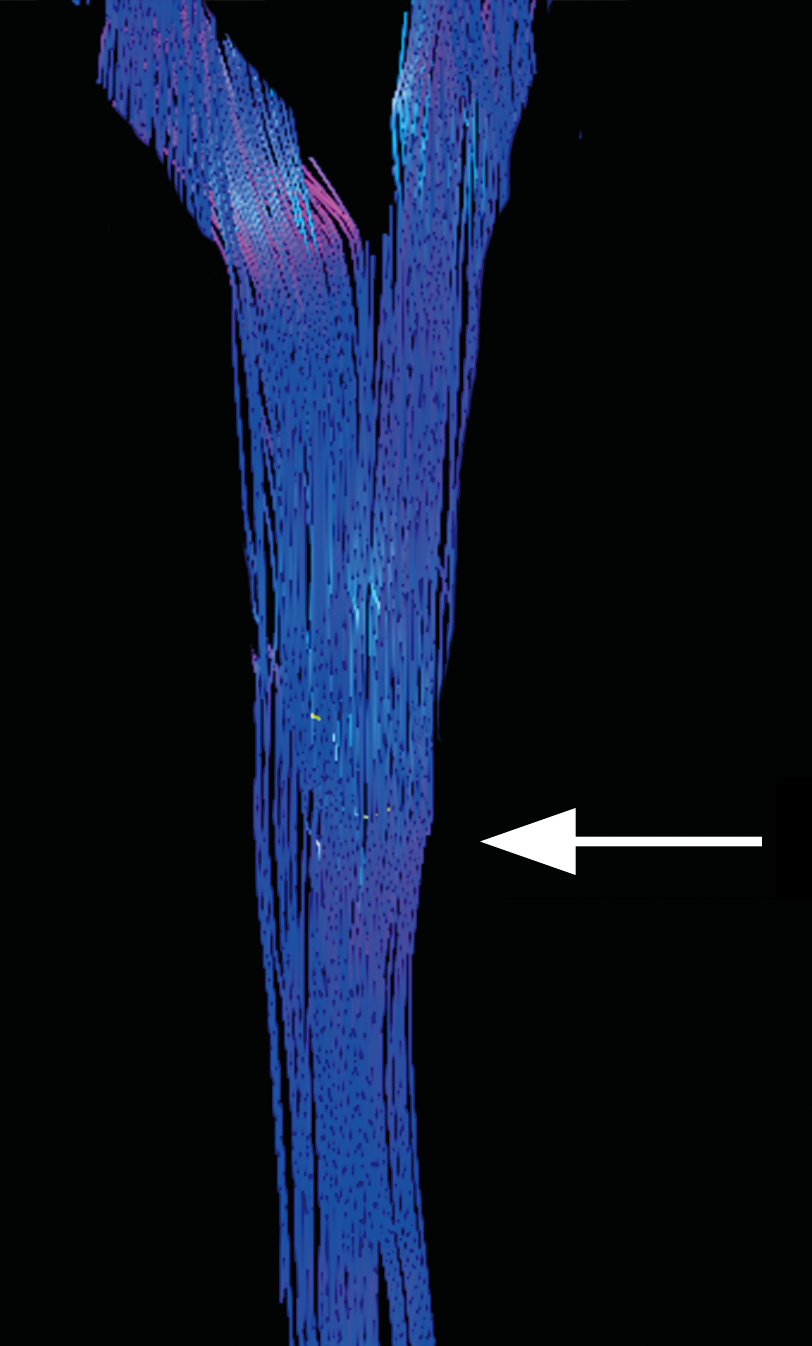
As a baseline for the acquired data, standard DTI processing was first examined. As seen in Figure 1, the crossing fibers of the pyramidal decussation are not accurately resolved in Subject 1. The crossing fibers were also unable to be seen utilizing standard DTI processing in Subject 2 or Subject 3. The tractography reveals the typical problems associated with standard DTI as the CST erroneously appears to travel straight through the medulla without decussating.
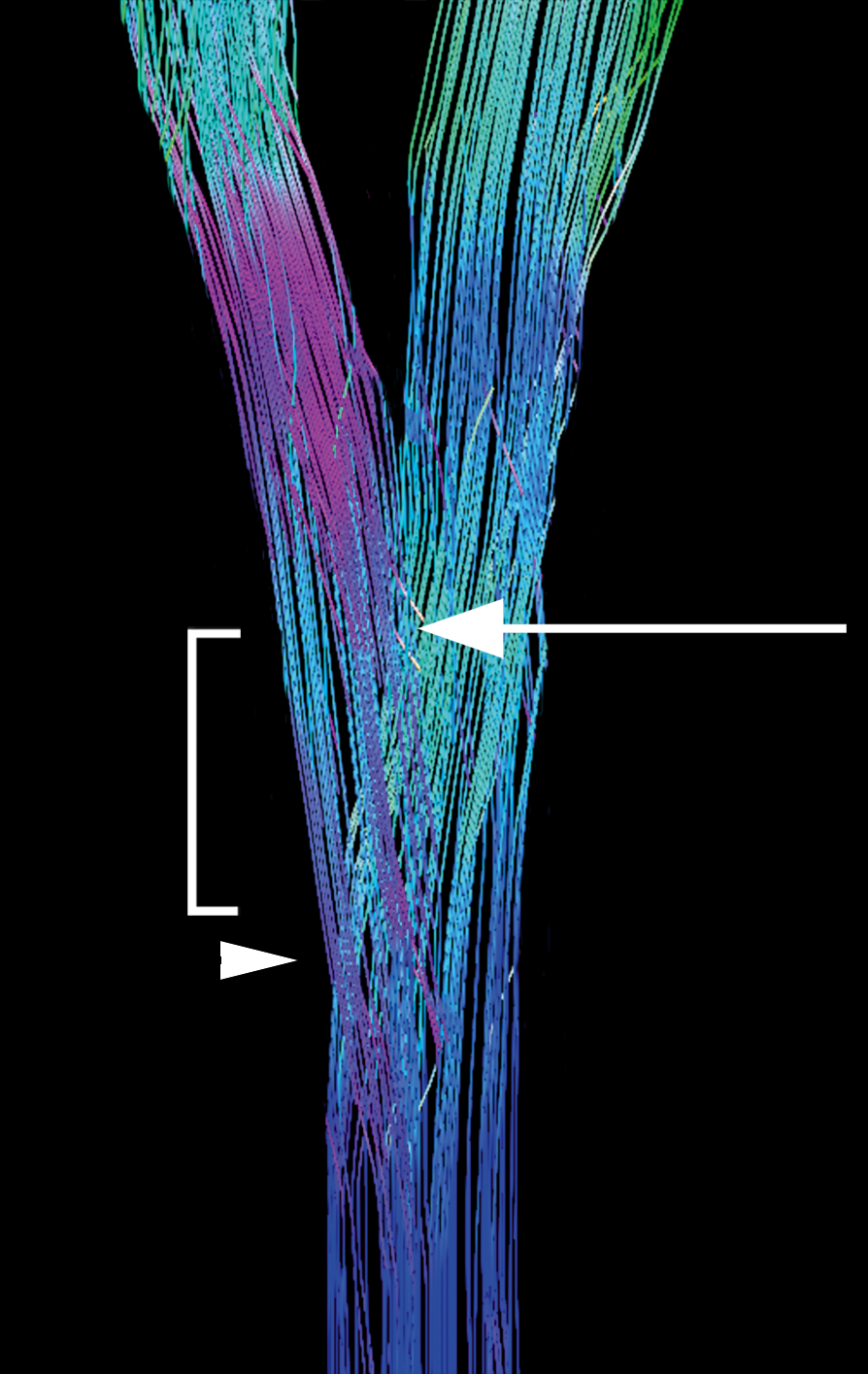
Subject 1 was then processed using GQI with diffusion decomposition. Substantial differences were seen in the ability to resolve the crossing fibers of interest. The pyramidal decussation is reliably identified in the lower medulla (Figure 2). The first crossing fibers occur within 5 mm of the obex and extend across multiple levels. A small percentage of fibers continue straight through the decussation to form the anterior corticospinal tract, as well as joining the ipsilateral lateral corticospinal tract.
Identical findings are also seen in Subject 2 (Figure 4) after GQI processing with diffusion decomposition. Analysis of Subject 3 shows that the pyramidal decussation is not reliably appreciated even with GQI and diffusion decomposition (Figure 5).
We have shown that standard DTI post-processing techniques have proven inadequate to reliably demonstrate the crossing fibers of the pyramidal decussation. These findings are not unexpected given the known difficulty with visualization of crossing fibers with DTI [5] ). When GQI with diffusion decomposition was utilized, the pyramidal decussation was identified using both a 10-minute and a 17-minute acquisition at 3 Tesla using isotropic voxels and 48 diffusion directions. A 5.5-minute acquisition at 30 directions proved to be inadequate to visualize the decussating fibers even when processed with GQI and diffusion decomposition.
Results show that the cervicomedullary junction can be reliably seen utilizing clinically feasible DTI acquisition techniques. Quisling et al demonstrated the initial crossing fibers of the pyramidal decussation occurred within 5 mm of the obex in normal pathologic specimens [3]. Our initial results of in vivo DTI agree with previously published anatomic data. Utilizing GQI with diffusion decomposition, we were able to accurately depict these crossing fibers using a clinically feasible DTI acquisition. In our small study, the number of diffusion directions appears to play a larger role in visualizing the decussation than the number of acquisitions alone.
Accurately visualizing the decussation of the pyramids on clinical imaging protocols will now give researchers the tools necessary to study the in vivo effects of an abnormal cervicomedullary junction. The pathological implications of an abnormally positioned cervicomedullary junction in conditions such as Chiari malformation, as well as potential outcomes of intervention, can now be easily studied. Additionally, pyramidal decussation abnormalities in less common conditions such as Joubert syndrome and HGPS, can also be studied in vivo for the first time.
The corticospinal tract and pyramidal decussation can be reliably visualized by clinically feasible DTI acquisitions utilizing GQI with diffusion decomposition.
No portion of this paper has been previously presented or published elsewhere. No financial support or funding was provided for the study. The authors have no conflicts of interest. The study was exempted from full IRB review by meeting University of Florida IRB requirements for “Case Report.”
- Wolpert S, Scott R, Platenberg C, Runge V. The clinical significance of hindbrain herniation and deformity as shown on MR images of patients with Chiari II malformation. AJNR Am J Neuroradiol. 1988;9:1075-8 pubmed
- Yamazaki Y, Tachibana S, Takano M, Fujii K. Clinical and neuroimaging features of Chiari type I malformations with and without associated syringomyelia. Neurol Med Chir (Tokyo). 1998;38:541-6; discussion 546-7 pubmed
- Quisling R, Quisling S, Mickle J. Obex/nucleus gracilis position: its role as a marker for the cervicomedullary junction. Pediatr Neurosurg. 1993;19:143-50 pubmed
- Vulliemoz S, Raineteau O, Jabaudon D. Reaching beyond the midline: why are human brains cross wired?. Lancet Neurol. 2005;4:87-99 pubmed
- Wiegell M, Larsson H, Wedeen V. Fiber crossing in human brain depicted with diffusion tensor MR imaging. Radiology. 2000;217:897-903 pubmed
- Tuch D, Reese T, Wiegell M, Makris N, Belliveau J, Wedeen V. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med. 2002;48:577-82 pubmed
- Alexander D, Barker G, Arridge S. Detection and modeling of non-Gaussian apparent diffusion coefficient profiles in human brain data. Magn Reson Med. 2002;48:331-40 pubmed
- Yeh F, Tseng W. Sparse solution of fiber orientation distribution function by diffusion decomposition. PLoS ONE. 2013;8:e75747 pubmed
- Yeh F, Wedeen V, Tseng W. Generalized q-sampling imaging. IEEE Trans Med Imaging. 2010;29:1626-35 pubmed
- King M, Houseman J, Roussel S, van Bruggen N, Williams S, Gadian D. q-Space imaging of the brain. Magn Reson Med. 1994;32:707-13 pubmed
- Wedeen V, Wang R, Schmahmann J, Benner T, Tseng W, Dai G, et al. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage. 2008;41:1267-77 pubmed
- Aggarwal M, Zhang J, Pletnikova O, Crain B, Troncoso J, Mori S. Feasibility of creating a high-resolution 3D diffusion tensor imaging based atlas of the human brainstem: a case study at 11.7 T. Neuroimage. 2013;74:117-27 pubmed
